File:Fpls-05-00179-g001 (14444289953).jpg

Original file (627 × 825 pixels, file size: 360 KB, MIME type: image/jpeg)
Captions
Captions
Summary[edit]
| DescriptionFpls-05-00179-g001 (14444289953).jpg |
Source: Front. Plant Sci., 01 May 2014 | doi:10.3389/fpls.2014.00179 Members of the MYBMIXTA-like transcription factors may orchestrate the initiation of fiber development in cotton seeds Frank Bedon, Lisa Ziolkowski, Sally A. Walford, Elizabeth S. Dennis and Danny J. Llewellyn Figure 1. Phylogenetic analysis, chromosomal location, gene expression of the cotton MML genes in wild type and transgenic G. hirsutum, and transactivation of the GhMML3 promoter in cotton protoplasts. (A) Phylogenetic analysis of the G. raimondii (Gr, filled diamonds) MML proteins and their putative G. hirsutum (Gh, empty diamonds) D-genome homologs. Four clades (Sg9-1 to 4) are indicated by shading. The rooted Neighbour–Joining tree was obtained in MEGA 5.0 with Clustal W alignment using the full length amino acid sequences (details in Supplementary data 1). Arabidopsis thaliana (At) MYB3R4 and Populus tremula × P. Tremuloides (Ptt) MYBR3 are R1R2R3-MYBs used as outgroups and the other AtMYB and Anthirrinum majus (Am) sequences are landmarks of Subgroup 9. (B) Schematic of the chromosomal distribution of the 10 GrMML genes indicated by the filled triangles, while the unfilled triangle represents a fragment of a MML gene. The directions of the triangle indicate the coding strand of the transcripts and the numbers under the triangle the particular GrMML gene. Adjacent triangles are tandemly arranged genes. (Chromosomes are only approximately to scale). The heat maps visualize the transcript level differences between the G. hirsutum homologues of the GrMML genes in: (C) cotton fibers, ovules and selected other plant organs, (D) three dissected tissues (OI, II and N) from wild type (G. hirsutum) ovules collected the day of anthesis [DPA(0)], (E) dissected OI of wild type ovules collected from 4 days before anthesis [DPA(−4)] to 2 days after anthesis [DPA(+2)], (F) OI dissected from G. hirsutum ovules silenced by RNAi for GhMYB25Like (ie., MYBML3) and GhMYB25 (i.e., MYBML7) and their respective controls (ie., null segregant plants) collected at DPA(−4), (−2), (0), and (+2) (F). Heat maps were made using Expander software based on gene expression relative to the cotton ubiquitin gene and normalized for each MML gene and separate experiment (details in Supplementary data 2). Primers used detect both the A- and D-genome homoeologues of each MML gene. (G) Transactivation assay of the GhMML3/GhMYB25 promoter-Luciferase reporter by GhMYB25Like and/or GhHD-1 in cotton cotyledon protoplasts (details in Supplementary data 3). |
| Date | |
| Source | fpls-05-00179-g001 |
| Author | Phylogeny Figures |
Licensing[edit]
- You are free:
- to share – to copy, distribute and transmit the work
- to remix – to adapt the work
- Under the following conditions:
- attribution – You must give appropriate credit, provide a link to the license, and indicate if changes were made. You may do so in any reasonable manner, but not in any way that suggests the licensor endorses you or your use.
| This image was originally posted to Flickr by phylofigures at https://flickr.com/photos/123636286@N02/14444289953. It was reviewed on 19 December 2020 by FlickreviewR 2 and was confirmed to be licensed under the terms of the cc-by-2.0. |
19 December 2020
File history
Click on a date/time to view the file as it appeared at that time.
| Date/Time | Thumbnail | Dimensions | User | Comment | |
|---|---|---|---|---|---|
| current | 14:11, 19 December 2020 | 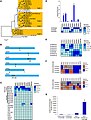 | 627 × 825 (360 KB) | Eyes Roger (talk | contribs) | Transferred from Flickr via #flickr2commons |
You cannot overwrite this file.
File usage on Commons
There are no pages that use this file.
Metadata
This file contains additional information such as Exif metadata which may have been added by the digital camera, scanner, or software program used to create or digitize it. If the file has been modified from its original state, some details such as the timestamp may not fully reflect those of the original file. The timestamp is only as accurate as the clock in the camera, and it may be completely wrong.
| Date and time of data generation | 07:38, 16 April 2014 |
|---|---|
| Width | 720 px |
| Height | 1,040 px |
| Compression scheme | LZW |
| Orientation | Normal |
| Number of components | 4 |
| Horizontal resolution | 72 dpi |
| Vertical resolution | 72 dpi |
| Data arrangement | chunky format |
| Software used | Shotwell 0.18.0 |
| File change date and time | 07:38, 16 April 2014 |
| Date and time of digitizing | 07:38, 16 April 2014 |
| Color space | Uncalibrated |
| IIM version | 12,981 |
| Keywords | Frontiers |
| Image width | 627 px |
| Image height | 825 px |
| Date metadata was last modified | 12:40, 29 April 2014 |
| Rating (out of 5) | 0 |
| Unique ID of original document | xmp.did:4617D697FDC5E311A946F6F564A9097C |